
Hi, welcome to my personal web site! I was born on 1984 in Hunan Province, which locates at the middle south part of the mainland China. Since 2010 I started my six years PhD training in the Institute of Genetics and Developmental Biology major in Genetics (bioinformatics direction). I studied how heterochromatin and heterochromatin gene evolution in a comparative genomics way. After bioinformatic traing, I continuied PostDoc in (epigentical) gene expression regulatory in Reproduction biology (embryo and placenta). I devoted myself as a computational biologist with more than fourteen years experience in the fields of comparative genomics, epigenetics, single cell/nucleus sequenencing. I am good at cooperation with biologist and eager to handle standalone project (both story driven and methodology driven).
Based on emphasis of bioinformatic methodology (both wet and dry), I mainly devote myself on the following research aspects:
Biology story driven:
(1) Reproduction Biology. Extraembrionic (placenta) and embryoinic cell lineage commitment; (2) Cardic development. Cardic lineage development and gene regulation in mammalian heart. Abnormal gene regulation during heart disease; (3) Neurology development. Neurologic tissue lineage commitment and gene regulation.
Methodology driven:
(1) Single cell/nucleus multiomic sequencing. scRNA-seq, snRNA+snATAC sequencing, scRNA-seq + CITE-seq ...; (2) Spatial single cell sequencing. Sequencing-based: Steroseq (BGI), Visum (10X), probe-based: MERFISH ...; (3) Single cell/nucleus multiomic-based gene regulation network construction.
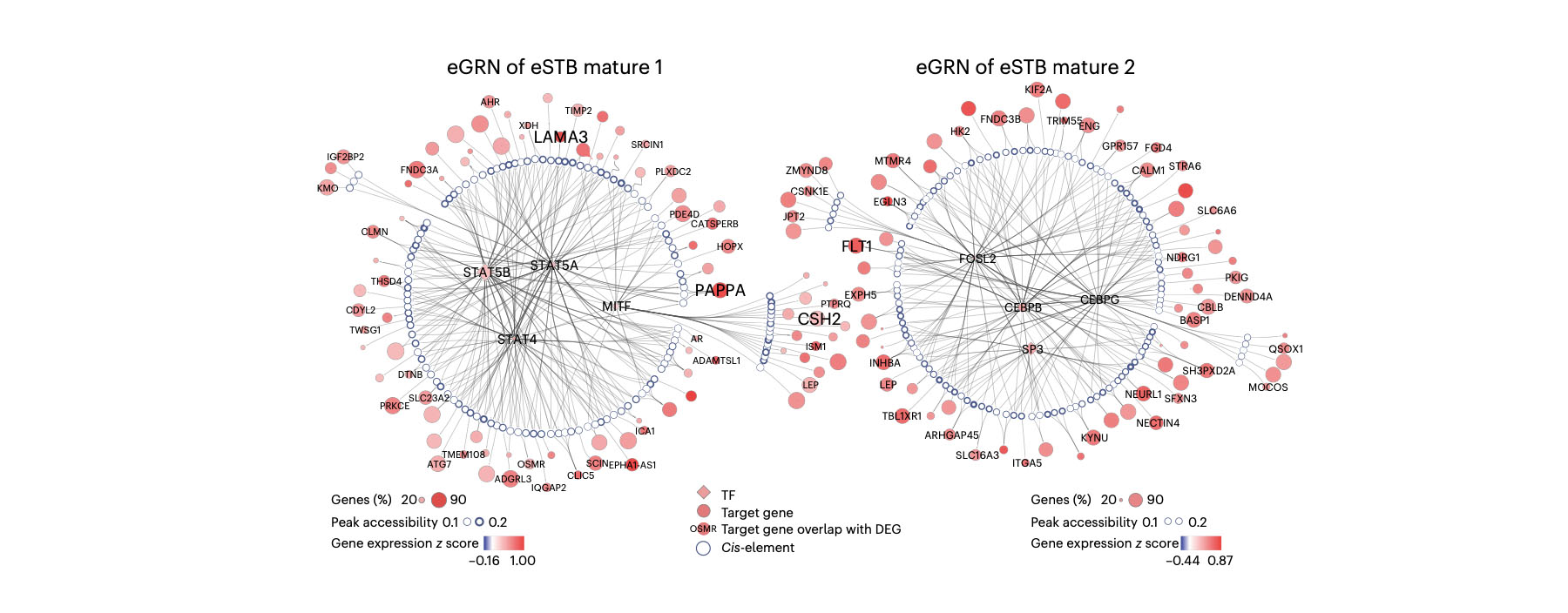
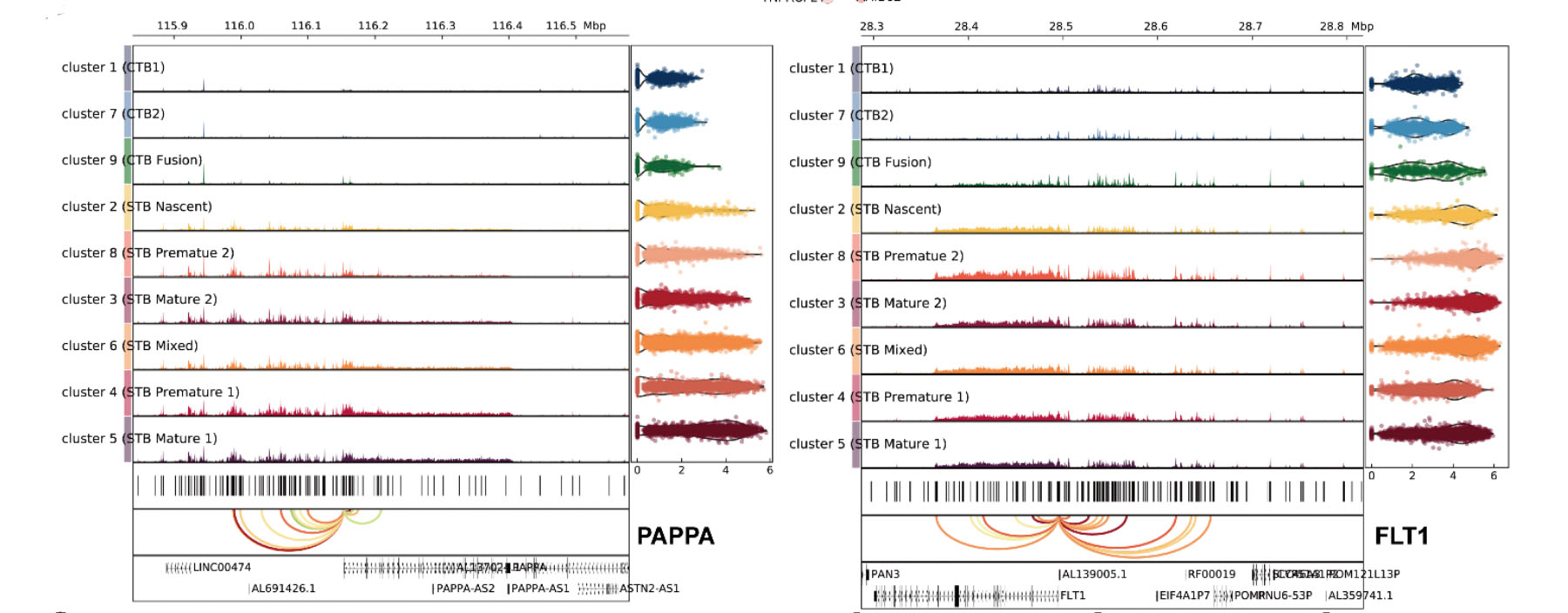
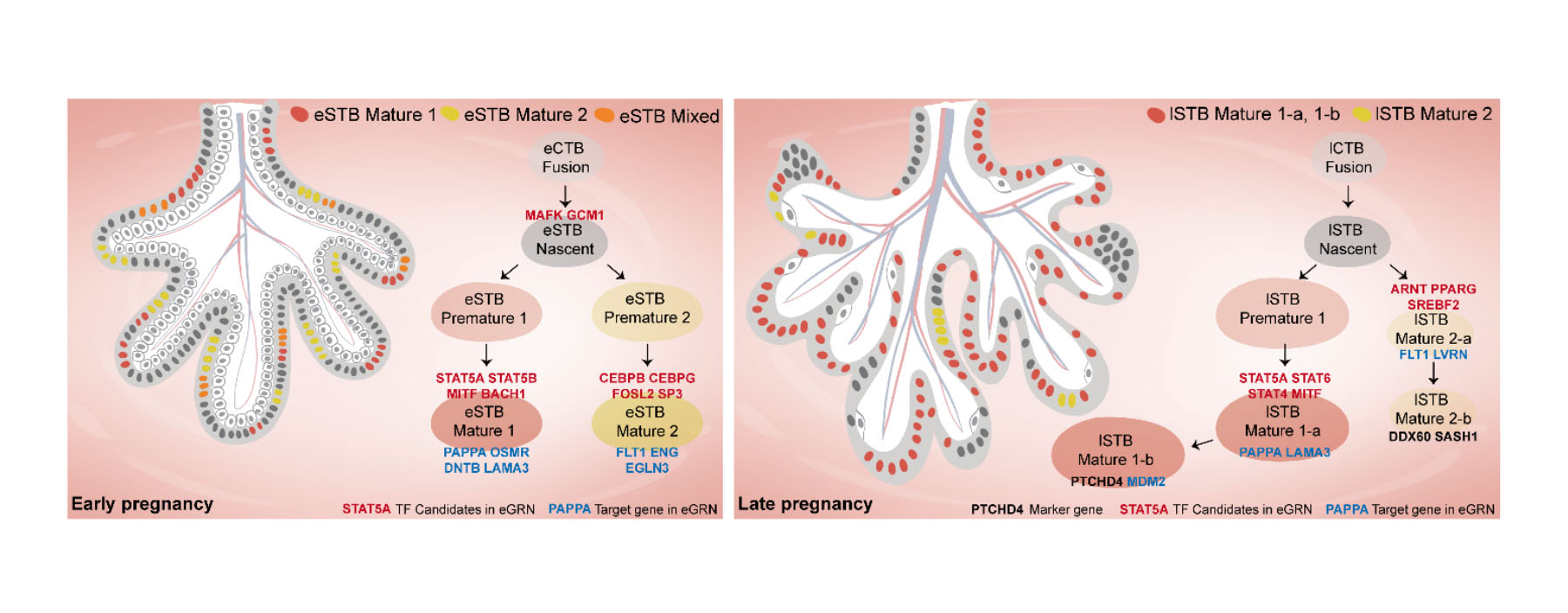
1. Wang M#, Liu Y#, Sun R#, Liu F#, Li J, Yan L, Zhang J, Xie X, Li D, Wang Y, Li S, Zhu X, Li R*, Lu F*, Xiao Z*, Wang H*. Single-nucleus multi-omic profiling of human placental syncytiotrophoblasts identifies cellular trajectories during pregnancy. Nat Genet (2024) 56, 294-305. (Article)

Syncytiotrophoblast (STB) differentiation study.
Abstract: The human placenta has a vital role in ensuring a successful pregnancy. Despite the growing body of knowledge about its cellular compositions and functions,
there has been limited research on the heterogeneity of the billions of nuclei within the syncytiotrophoblast (STB), a multinucleated entity primarily
responsible for placental function. Here we conducted integrated single-nucleus RNA sequencing and single-nucleus ATAC sequencing analyses of human placentas
from early and late pregnancy. Our findings demonstrate the dynamic heterogeneity and developmental trajectories of STB nuclei and their correspondence with human
trophoblast stem cell (hTSC)-derived STB. Furthermore, we identified transcription factors associated with diverse STB nuclear lineages through their gene regulatory
networks and experimentally confirmed their function in hTSC and trophoblast organoid-derived STBs. Together, our data provide insights into the heterogeneity of human
STB and represent a valuable resource for interpreting associated pregnancy complications.
PMID: 3826760
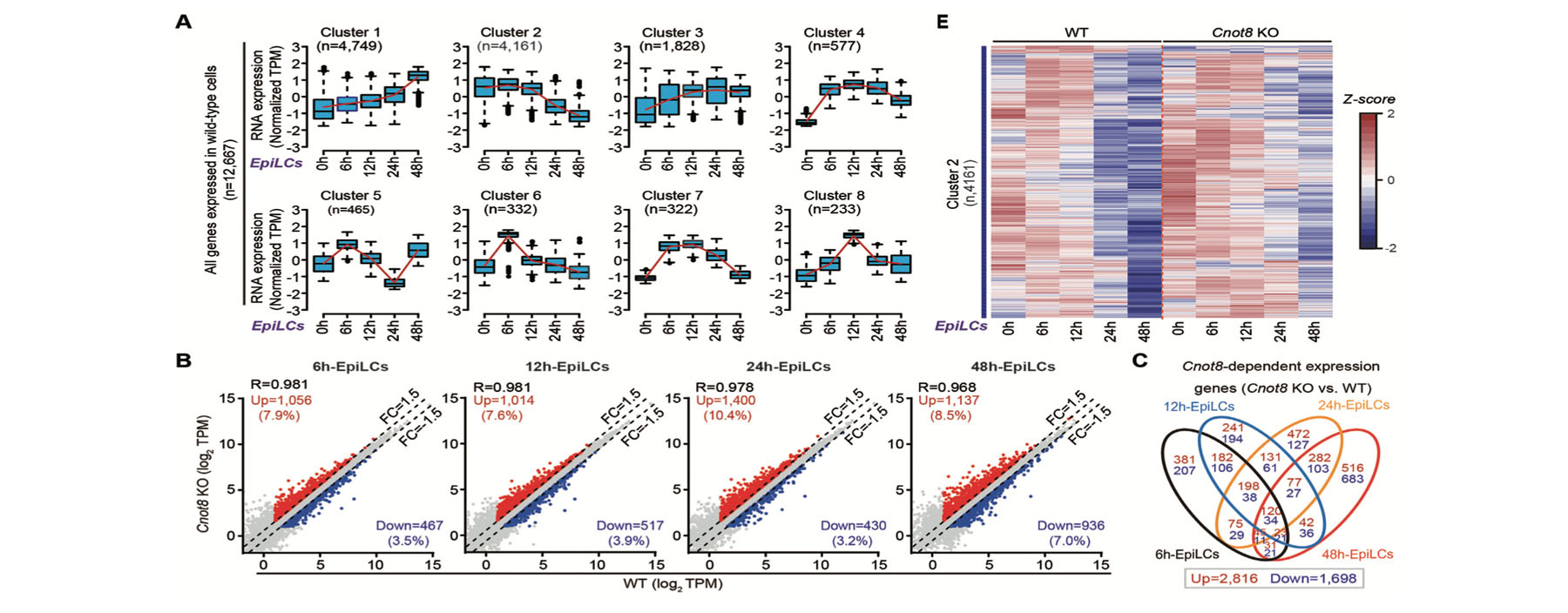
2. Quan Y#, Wang M#, Xu C, Wang X, Qin D, Lin Y, Lu X, Lu F*, Li L*. Cnot8 eliminates naive regulation networks and is essential for naive-to-formative pluripotency transition. Nucleic Acids Res (2022) 50(8):4414-4435.

Cnot8 driven embryonic pluripotency transition.
Abstract: Mammalian early epiblasts at different phases are characterized by naïve, formative, and primed pluripotency states, involving extensive transcriptome changes.
Here, we report that deadenylase Cnot8 of Ccr4-Not complex plays essential roles during the transition from naïve to formative state.
Knock out (KO) Cnot8 resulted in early embryonic lethality in mice, but Cnot8 KO embryonic stem cells (ESCs) could be established.
Compared with the cells differentiated from normal ESCs, Cnot8 KO cells highly expressed a great many genes during their differentiation into the formative state,
including several hundred naïve-like genes enriched in lipid metabolic process and gene expression regulation that may form the naïve regulation networks.
Knockdown expression of the selected genes of naïve regulation networks partially rescued the differentiation defects of Cnot8 KO ESCs. Cnot8 depletion led to
the deadenylation defects of its targets, increasing their poly(A) tail lengths and half-life, eventually elevating their expression levels. We further found that
Cnot8 was involved in the clearance of targets through its deadenylase activity and the binding of Ccr4-Not complex, as well as the interacting with Tob1 and Pabpc1.
Our results suggest that Cnot8 eliminates naïve regulation networks through mRNA clearance, and is essential for naïve-to-formative pluripotency transition.
PMID: 35390160
3. Wang M#, Zhang Y#, Lin ZS*, Ye XG, Yuan YP, Ma W, Xin ZY*. Development of EST-PCR markers for Thinopyrum intermedium chromosome 2Ai#2 and their application in characterization of novel wheat-grass recombinants. Theor Appl Genet (2010) 121(7): 1369-1380.

A comparative genomics study in earlier year.
PMID: 20585749
4. Wang Y#, Wu H#, Jiang X#, Jia L#, Wang M, Rong Y, Chen S, Wang Y, Xiao Z*, Liang X*, Wang H*. LMNA Determines Nuclear Morphology During Syncytialization of Human Trophoblast Stem Cells. Front Cell Dev Biol. (2022) 10:836390.
5. Jiang X#, Zhai J#, Xiao Z#, Wu X#, Zhang D#, Wan H, Xu Y, Qi L, Wang M, Yu D, Liu Y, Wu H, Sun R, Xia S, Yu K, Guo J, Wang H*. Identifying a dynamic transcriptomic landscape of the cynomolgus macaque placenta spanning during pregnancy at single-cell resolution. Dev Cell. (2023) 58:806-821.
6. Wu X#, Zhao W, Wu H, Zhang Q, Wang Y, Yu K, Zhai J, Mo F, Wang M, Li S, Zhu X, Liang X, Hu B, Liu G, Wu J, Wang H, Guo F, Yu L. An aggregation of human embryonic and trophoblast stem cells reveals the role of trophectoderm on epiblast differentiation. Cell Prolif. (2023) 56, e13492.
7. Chen J, Huang Q, Gao D, Wang J, Lang Y, Liu T, Li B, Bai Z, Luis Goicoechea J, Liang C .. Wang M, .. Wing RA, Chen M. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nature communications (2013) 4: 1595.
8. Bai Z, Chen J, Liao Y, Wang M, Liu R, Ge S, Wing RA, Chen M. The impact and origin of copy number variations in the Oryza species. BMC genomics (2016)17: 261.
9. Wei G, Tian P, Zhang F, Qin H, Miao H, Chen Q, Hu Z, Cao L,Wang M, Gu X et al. Integrative Analyses of Nontargeted Volatile Profiling and Transcriptome Data Provide Molecular Insight into VOC Diversity in Cucumber Plants (Cucumis sativus). Plant physiology (2016) 172(1): 603-618.
10. Liao Y, Zhang X, Li B, Liu T, Chen J, Bai Z, Wang M, Shi J, Walling JG, Wing RA et al. Comparison of Oryza sativa and Oryza brachyantha Genomes Reveals Selection-Driven Gene Escape from the Centromeric Regions. The Plant cell (2018) 30(8): 1729-1744.
11. Tian P, Zhang X, Xia R, Liu Y, Wang M, Li B, Liu T, Shi J, Wing R, Meyers B, Chen M. Evolution and diversification of reproductive phased small interfering RNAs in Oryza species. NEW PHYTOLOGIST (2021) 229(5): 2970-2983
12. Wang M#, Zou H#, Lin Z, Wu Y, Chen X, Yuan Y. Expressed sequence tag-PCR markers for identification of alien barley chromosome 2H in wheat. Genetics and molecular research (2012) 11(3): 3452-3463.
A critical cook.